This page is designed to explain how outerbase can
facilitate fast inference with smart modeling choices.
The potential benefits grow as the sample size grows. We use a sample
size of 500 here in the spirit of running quickly. The
point will be obvious, but more dramatic results can be had by
increasing the sample size.
sampsize = 500
d = 8
x = matrix(runif(sampsize*d),ncol=d)
y = obtest_borehole8d(x)First setup an outermod object.
om = new(outermod)
setcovfs(om, rep("mat25pow",8))
knotlist = list();
for(k in 1:d) knotlist[[k]] = seq(0.01,1,by=0.025)
setknot(om, knotlist) #40 knot point for each dimMore data should mean more basis functions. So we will choose
250 terms for our feature space approximation.
p = 250
terms = om$selectterms(p)Different models
To begin, lets use ?loglik_std to represent our slow
approach.
loglik_slow = new(loglik_std, om, terms, y, x)
logpr_slow = new(logpr_gauss, om, terms)
logpdf_slow = new(lpdfvec, loglik_slow, logpr_slow)logpdf_slow can be optimized using
lpdf$optnewton.
logpdf_slow$optnewton()Newton’s method involves solving a linear system, thus it takes one step, but is expensive.
?loglik_gauss is a lpdf model designed for
speed. It is a nice comparison because loglik_gauss uses
the same model as loglik_std, with a few approximations for
speed.
loglik_fast = new(loglik_gauss, om, terms, y, x)
logpr_fast = new(logpr_gauss, om, terms)
logpdf_fast = new(lpdfvec, loglik_fast, logpr_fast)logpdf_fast will through an error if you try to use
optnewton. This is because it is written so that it never
builds a Hessian (hess in the code) matrix.
logpdf_fast$optnewton()
#> Error in eval(expr, envir, enclos): addition: incompatible matrix dimensions: 0x0 and 250x250It is instead suggested to use lpdf$optcg (conjugate
gradient) to optimize the coefficients in the fast version.
logpdf_fast$optcg(0.001, # tolerance
100) # max epochsAs an aside, omp speed ups are possible, but you need to
have correctly compiled with omp. One check is to call the
following.
ob = new(outerbase, om, x)
ob$nthreads
#> [1] 2If the answer is 1 but you have a multicore processor
(most modern processors), your installation might be incorrect.
You can manually set the number of threads for lpdf
objects.
logpdf_slow$setnthreads(4)
logpdf_fast$setnthreads(4)Timing
The main cost of fitting outerbase models is
hyperparameter optimization. The difference between
logpdf_slow and logpdf_fast will be apparent.
Let’s save starting points (since they share om) for
fairness.
parlist_slow = list(para = getpara(logpdf_slow), hyp = gethyp(om))
parlist_fast = list(para = getpara(logpdf_fast), hyp = gethyp(om))Test points will verify the predictions are equally good with either model, the only difference is speed.
xtest = matrix(runif(1000*d),ncol=d) #prediction points
ytest = obtest_borehole8d(xtest)We will use the unsophisticated proc.time to do some
quick timing comparisons.
Comparison of results
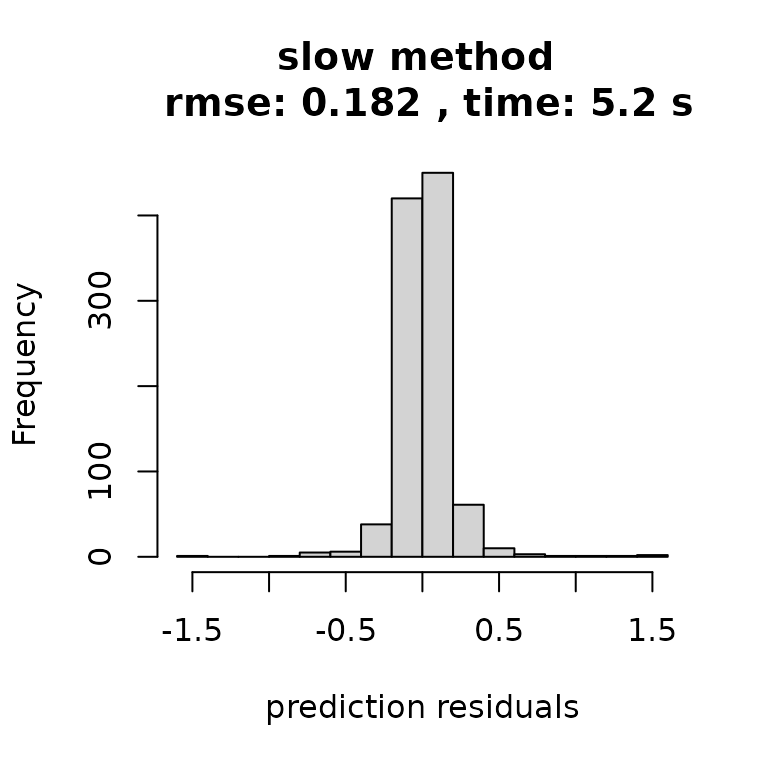
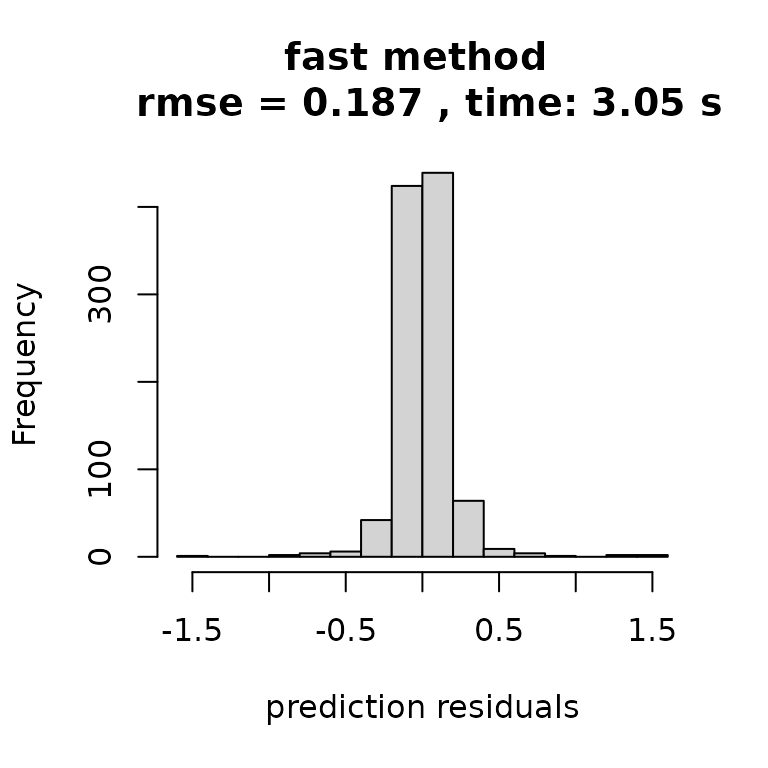
And simply plotting the results tells the story: faster inference with no discernible drop off in quality. Note there are serious approximations here, but the approximations just have a negligible effect.
rmse_slow = sqrt(mean((ytest-yhat_slow)^2))
hist((ytest-yhat_slow), main=paste("slow method \n rmse:",
round(rmse_slow,3),
", time:",
round(t_slow[3],2),'s'),
xlab = "prediction residuals")
rmse_fast = sqrt(mean((ytest-yhat_fast)^2))
hist((ytest-yhat_fast), main=paste("fast method \n rmse =",
round(rmse_fast,3),
", time:",
round(t_fast[3],2),'s'),
xlab = "prediction residuals")